
Dear Reader,
With our 14th NFDI4Microbiota newsletter, we would like to share news about conferences, training courses, services and publications.
If you are interested in other topics we should cover, please let us know. We are happy to hear from you!
And now: Enjoy reading the newsletter!
Consortium News
A foreword from the NFDI4Microbiota Board of Directors


As we begin 2026, we take a moment to look back on a remarkable year for NFDI4Microbiota. The year 2025 was marked by several achievements, and we are deeply grateful for the contributions by the supporting scholarly societies, participants and partners, which made it all possible. Reflecting on the past year, we are especially proud to have passed our mid-term evaluation in January 2025 with flying colors, earning high praise from the reviewers. This milestone set the tone for a year of continued success. Our key services, launched as part of our program, not only met but exceeded expectations, attracting impressive user numbers throughout 2025. These services are now established as supporting tools for FAIR and open research in microbiology in Germany, Europe and beyond. Alongside them, our training and outreach activities, the service helpdesk, the knowledge base, and our use cases have all flourished - with even further developments on the horizon for the final year of our first funding period. We also strengthened our collaborations with other NFDI consortia in the life sciences and expanded our activities within the BioMed and Biodata interest groups, to generate solutions for the broader scientific community. Furthermore, we contributed in the discussions regarding the future structure and governance of NFDI always with the vision of “One NFDI” in mind. A particular highlight was our annual conference in Cologne, which brought together numerous community members for learning, networking, and meaningful exchange. As we look ahead, we are awaiting the outcome of our second proposal, hoping to continue our mission into the future. Until then, we wish you all a year filled with success, health, and happiness in 2026!
Defense of the NFDI4Microbiota second funding phase proposal

After successfully submitting our renewal proposal for the second funding phase, we presented and defended it before the DFG reviewers in October 2025. The discussion was highly constructive and offered valuable insights into both our past achievements and our plans for the coming years. If the evaluation is positive, our work will continue from October 2026 onward. The consortium sincerely thanks the reviewers and the DFG for their time and thoughtful feedback, as well as NFDI e.V. for its continuous support. We are excited about the road ahead!
BioMed Interest Group strengthens collaboration on the path to One NFDI
.jpg)
The NFDI BioMed interest group – jointly formed by GHGA, NFDI4BIOIMAGE, NFDI4Health, NFDI4Immuno and NFDI4Microbiota – aims at advancing biomedical data management across NFDI. The workshop was held from December 2-4, 2025 at ZB MED Cologne to define next steps for continued collaboration. During the strategy workshop, the consortium speakers explored how BioMed can contribute to a more unified structure within the broader NFDI, and developed a shared definition of services. In the outreach session, several action points for future collaboration were refined. The existing services and training catalogs are being consolidated to identify overlaps and synergies. Additionally, the user’s need for a shared metadata platform is being assessed through a survey. Finally, existing offers for helpdesk and websites are being enhanced by referring to the other consortia. The workshop further strengthened connections within the BioMed interest group and was a step towards the vision of One NFDI.
Community engagement – calls, events and conferences
Joint NFDI4Microbiota Annual Conference 2026

Save the date for the 5th NFDI4Microbiota Annual Conference on September 15 -17, 2026! This year, the conference will be a joint event organized together with other life science consortia of the NFDI biodata interest group, highlighting the full spectrum of NFDI biodata services. Stay tuned—more details will be announced soon!
Meet us at the VAAM and DGE 2026
We are excited to meet our research community in March 2026 at the Annual Conference of the Association for General and Applied Microbiology (VAAM) and at the Conference of the German Nutrition Society (DGE), which will focus on nutrition and the microbiome this year. NFDI4Microbiota will have a booth at both events and will also host a lunch symposium at the VAAM 2026. Join us to learn more about FAIR-compliant research data management and analysis in microbiology!
Impressions from the NFDI4Microbiota Annual Conference 2025

The fourth NFDI4Microbiota Annual Conference took place from September 30 to October 2, 2026, at ZB MED in Cologne. We were delighted to welcome around 50 external participants from our community, including PIs, data stewards, bioinformaticians, and early career researchers. Several attendees also joined our Ambassador Program, which aims to connect and train scientists across the microbiology community.

Across nine hands-on workshops, we taught FAIR research data management and microbiological data analysis and discussed how participants can apply these principles in their daily work. Many attendees highlighted that they especially appreciated the introductory, easy-to-follow workshop formats, the hands-on elements, and the opportunity to reflect on their own research practices. The poster session proved to be a lively networking space, and the diversity of topics and participants (from wet lab to dry lab to data stewardship) was frequently mentioned as a major strength of the event.
Our three keynote speakers — Samantha Pearman-Kanza, Alexandre Smirnov, and Ruth Schmitz-Streit — provided inspiring insights into implementing Electronic Lab Notebooks, practical, real-world examples of FAIR implementation, and current trends in microbiology research. A panel discussion on the intersection of AI and wet-lab science also drew great interest. Overall, the conference was described as “refreshing,” “friendly,” and “inspiring,” with participants emphasizing the importance of discussing FAIR practices across all career stages, from students to PIs. We thank everyone for their contributions, ideas, and enthusiasm, and we look forward to continuing this journey together.

Recap on recent workshops and conferences
The 4th biannual NFDI4Microbiota Knowledge Base Sprint was held as a lunch-to-lunch hybrid event, based in Jena, on November 24-25, 2025. There were 15 participants across the two days, whose contributions resulted in 48 git commits and 29 files updated. We had nice discussions and coffee breaks together online, and in person shared a tour around the new office building in Jena. The knowledge base is a collection of information and references with relevance for the microbiology community in terms of research data management, data analysis, FAIR principles, standards, and much more.
On October 7, 2025, researchers met at the Center for Molecular Biosciences in Kiel for the Biometadata-05 Workshop, “How to describe biological data? A primer to a FAIR approach for now and the future.” Organized by NFDI4Microbiota and hosted by miTarget, the workshop provided a practical introduction to creating FAIR-compliant microbiome metadata. Participants learned how to use controlled vocabularies and ontologies to describe biological datasets, practiced annotating metadata for their own projects, and discussed common challenges such as mapping experimental details to standard fields or documenting missing information. Well-structured metadata is essential for reproducible and reusable microbiome research, and the workshop helped equip attendees with the skills needed to prepare and share high-quality metadata. Looking ahead, participants are encouraged to apply these practices in their ongoing projects and future data submissions. Find more impressions from the workshop here.

SFBs and research institutes interested in FAIR biometadata training in 2026 are welcome to contact us via the NFDI4Microbiota helpdesk.
Refinement of the NFDI4Microbiota Ambassador Program
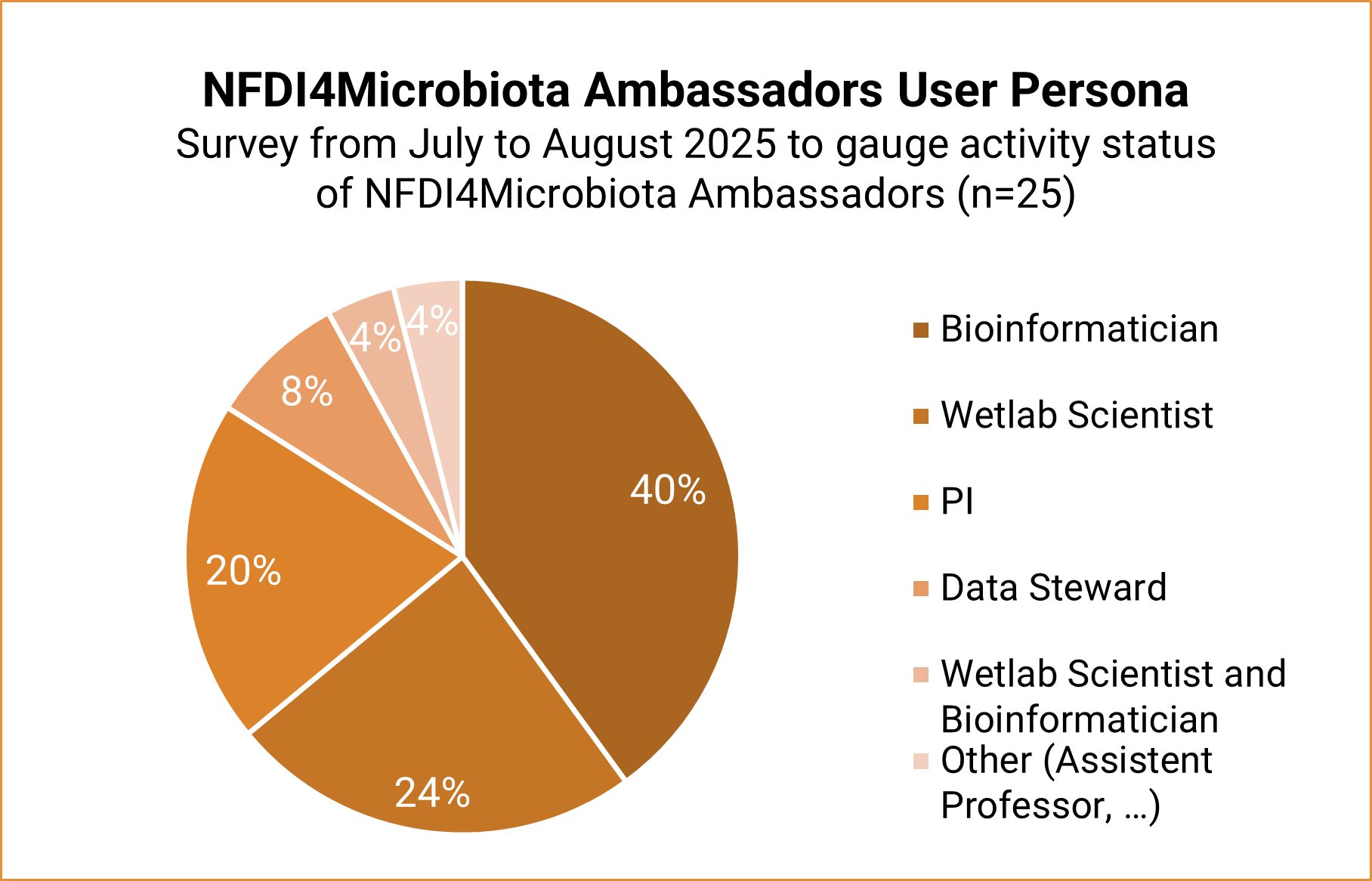
The NFDI4Microbiota Ambassador Program helps us establish direct connections with local institutes and research initiatives within the microbial research community. This network is essential for driving the cultural change in microbial research data management, as our Ambassadors actively promote our mission and inform their local communities about our services including training opportunities and individual support via our helpdesk. At the same time, we greatly benefit from the valuable feedback they provide, which helps us continuously improve our services. To further strengthen our collaboration, we will restructure our bi-monthly Ambassador meetings to foster a more interactive and informal environment and to address topics relevant to the different user personas illustrated in the graphic and described on our website.
You can find more information about the Ambassador Program on our website or register directly here.
Take a break and join our Coffee Talks

The Coffee Talk meetings provide an opportunity to learn more about NFDI4Microbiota’s services and mission from within and around the NFD4Microbiota community. These bimonthly meetings take place online and are open to all who are interested in staying informed about current developments and topics related to data in microbiology research. Get your updates and more information on the schedule and registration information here.
Services
To support the microbial research community in data management and analysis, the NFDI4Microbiota consortium has established a range of dedicated services. Visit our persona webpage to explore which services are available for you as a wet-lab scientist, bioinformatician, data steward, or PI, or find an overview of all our services here.
Protologger is back online
Protologger — a central tool of the NFDI4Microbiota use case “TAXA” that automates bacterial protologue generation to ensure standardized taxonomic descriptions — is now fully accessible again. We apologize for any inconveniences this has caused! Due to recent technical issues, users will need to create a new account to continue using the service. The issue has been resolved, and new monitoring measures are in place to prevent similar downtime in the future. Give it a try and let us know if you encounter any problems!
Two NFDI4Microbiota co-funded services recognized as official de.NBI services
We are pleased to announce that two services co-funded by NFDI4Microbiota have recently been acknowledged as official services within the de.NBI network: CloWM and the MdoA tool box. This recognition is an important milestone. Inclusion in the de.NBI service portfolio is not automatic, it follows a strict evaluation process designed to ensure that all services meet high scientific, technical, and community standards. According to de.NBI’s assessment principles, services must remain dynamic, relevant, up-to-date, and aligned with the evolving needs of life science researchers. Continuous evaluation and maintenance ensure optimal performance and adherence to rigorous scientific criteria.
Ask your questions to our Helpdesk!

The NFDI4Microbiota Helpdesk is the point of contact for the microbiology research community for all questions related to microbial (omics) data and associated metadata. We welcome questions from anyone - students, researchers, data stewards and more - working with this type of data, regardless of organism (bacteria, archaea, eukaryotic microorganisms, viruses), environment (e.g. soil, plants, host-associated and water) or data type (e.g. nucleic acid sequences, functional genomics, image data). You can contact us either by filling in our contact form or by email: helpdesk@nfdi4microbiota.de.
Training announcements

Education and training are key goals within NFDI4Microbiota. To support our community, the consortium offers training opportunities on a variety of topics and across different disciplines. Explore our training offers here. If you like to stay up to date on training opportunities in research data management for the life sciences, we recommend to subscribe to this mailing list offered by NFDI and designed to keep professionals, students, and researchers in the biosciences regularly informed about upcoming training sessions, workshops, and continuing education opportunities.
Publications
StrainInfo—the central database for linked microbial strain identifiers
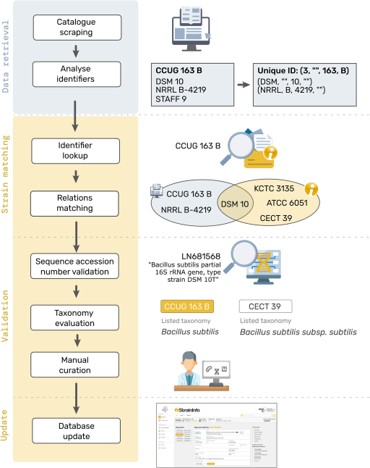
In microbiology, the communication and comparison of research findings and data is often complicated by the existence of a number of different designations and identifiers for the same strain. StrainInfo is a service developed by NFDI4Microbiota to provide a resolution of microbial strain identifiers by storing culture collection numbers, their relations, and deposition data. An article has been recently published and discusses the new StrainInfo database, developed to address the challenges of microbial strain identifiers across scientific literature and databases. By consolidating and standardizing strain data from multiple sources, StrainInfo improves the findability, reusability, and integration of published information related to strains. DOI: 10.1093/database/baaf059 .
Metalog: curated and harmonised contextual data for global metagenomics samples

Metagenomic sequencing enables the in-depth study of microbes and their functions in humans, animals, and the environment. While sequencing data is deposited in public databases, the associated contextual data is often not complete and needs to be retrieved from primary publications. This lack of access to sample-level metadata like clinical data or in situ observations impedes cross-study comparisons and metaanalyses. The authors of this article therefore created the Metalog database, a repository of manually curated metadata for metagenomics samples across the globe. It contains 80 423 samples from humans (including 66 527 of the gut microbiome), 10 744 animal samples, 5547 ocean water samples, and 23 455 samples from other environmental habitats such as soil, sediment, or fresh water. DOI: 10.1093/nar/gkaf1118 .
Fast and robust estimate of bacterial genus novelty using the percentage of conserved proteins with unique matches (POCPu)
Accurate taxonomic assignment of bacterial genomes is essential for identifying novel taxa and for stable classification to enable robust comparison between studies. In this study, the authors evaluated the commonly used Percentage of Conserved Proteins (POCP) and an improved version, POCPu which considers unique matches only, across 2.3 million proteome comparisons from 4,767 genomes. Using DIAMOND, POCPu run 20× faster than BLASTP and differentiates better within-genus from between-genera values than POCP, which improves bacterial genus assignment. DOI: 10.7717/peerj.20259 .
metaTraits: a large-scale integration of microbial phenotypic trait information

We are excited to announce the release of metaTraits, now published in Nucleic Acids Research. metaTraits harmonizes microbial phenotypic trait data, summarizing more than 140 traits across 2.2 million genomes in a single unified resource. By combining culturebased evidence with genome-based predictions, it integrates data silos and fills critical gaps for uncultured lineages. Initiated through an NFDI4Microbiota Flex Fund, metaTraits was developed by Daniel Podlesny and Chan Yeong Kim et al. from the Bork Group at EMBL Heidelberg, in collaboration with Lorenz Reimer and colleagues from the Leibniz Institute DSMZ. metaTraits provides workflows for annotating user-submitted genomes/MAGs and metagenomic profiles, and can be interactively explored at metaTraits. DOI: 10.1093/nar/gkaf1241 .